Example data to initialize scheduled events for a population of
1600 nodes and demonstrate the SISe3 model.
Usage
data(events_SISe3)Details
Example data to initialize scheduled events (see
SimInf_events) for a population of 1600 nodes
and demonstrate the SISe3 model. The dataset
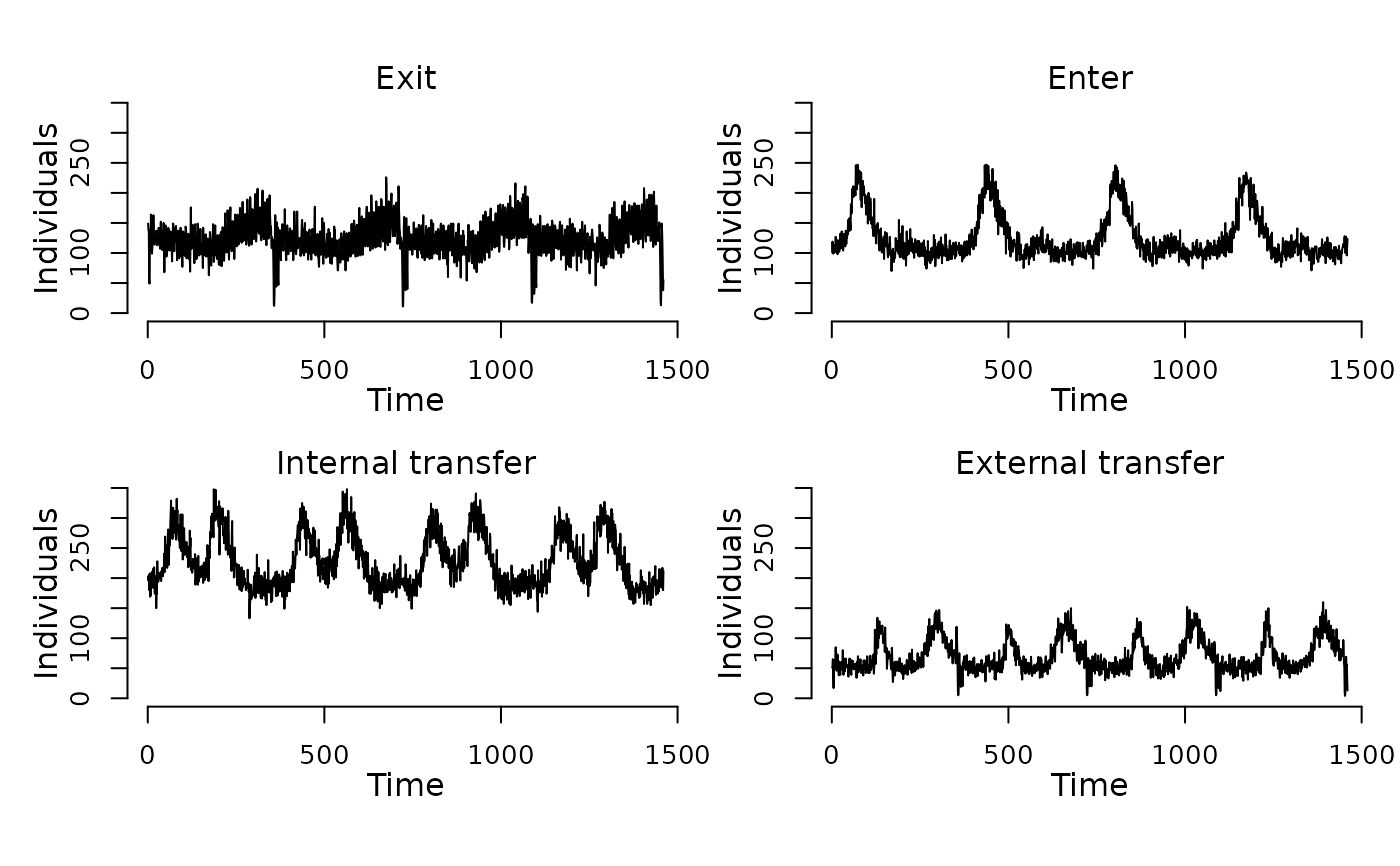
contains 783773 events for 1600 nodes distributed over 4 * 365
days. The events are divided into three types: ‘Exit’
events remove individuals from the population (n = 182535),
‘Enter’ events add individuals to the population (n =
182685), ‘Internal transfer’ events move individuals
between compartmens within one node e.g. ageing (n = 317081), and
‘External transfer’ events move individuals between nodes
in the population (n = 101472). The vignette contains a detailed
description of how scheduled events operate on a model.
Examples
## For reproducibility, call the set.seed() function and specify
## the number of threads to use. To use all available threads,
## remove the set_num_threads() call.
set.seed(123)
set_num_threads(1)
## Create an 'SISe3' model with 1600 nodes and initialize
## it to run over 4*365 days. Add one infected individual
## to the first node.
data("u0_SISe3", package = "SimInf")
data("events_SISe3", package = "SimInf")
u0_SISe3$I_1[1] <- 1
tspan <- seq(from = 1, to = 4*365, by = 1)
model <- SISe3(u0 = u0_SISe3, tspan = tspan, events = events_SISe3,
phi = rep(0, nrow(u0_SISe3)), upsilon_1 = 1.8e-2,
upsilon_2 = 1.8e-2, upsilon_3 = 1.8e-2,
gamma_1 = 0.1, gamma_2 = 0.1, gamma_3 = 0.1,
alpha = 1, beta_t1 = 1.0e-1, beta_t2 = 1.0e-1,
beta_t3 = 1.25e-1, beta_t4 = 1.25e-1, end_t1 = 91,
end_t2 = 182, end_t3 = 273, end_t4 = 365, epsilon = 0)
## Display the number of individuals affected by each event type
## per day.
plot(events(model))
 ## Run the model to generate a single stochastic trajectory.
result <- run(model)
## Summarize the trajectory. The summary includes the number of
## events by event type.
summary(result)
#> Model: SISe3
#> Number of nodes: 1600
#>
#> Transitions
#> -----------
#> S_1 -> upsilon_1*phi*S_1 -> I_1
#> I_1 -> gamma_1*I_1 -> S_1
#> S_2 -> upsilon_2*phi*S_2 -> I_2
#> I_2 -> gamma_2*I_2 -> S_2
#> S_3 -> upsilon_3*phi*S_3 -> I_3
#> I_3 -> gamma_3*I_3 -> S_3
#>
#> Global data
#> -----------
#> Parameter Value
#> upsilon_1 0.018
#> upsilon_2 0.018
#> upsilon_3 0.018
#> gamma_1 0.100
#> gamma_2 0.100
#> gamma_3 0.100
#> alpha 1.000
#> beta_t1 0.100
#> beta_t2 0.100
#> beta_t3 0.125
#> beta_t4 0.125
#> epsilon 0.000
#>
#> Local data
#> ----------
#> Parameter Value
#> end_t1 91
#> end_t2 182
#> end_t3 273
#> end_t4 365
#>
#> Scheduled events
#> ----------------
#> Exit: 182535
#> Enter: 182685
#> Internal transfer: 317081
#> External transfer: 101472
#>
#> Network summary
#> ---------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> Indegree: 40.0 57.0 62.0 62.1 68.0 90.0
#> Outdegree: 36.0 57.0 62.0 62.1 67.0 89.0
#>
#> Continuous state variables
#> --------------------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> phi 0.00e+00 0.00e+00 0.00e+00 8.92e-08 0.00e+00 1.88e-02
#>
#> Compartments
#> ------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> S_1 0.00e+00 7.00e+00 9.00e+00 9.23e+00 1.20e+01 3.00e+01
#> I_1 0.00e+00 0.00e+00 0.00e+00 1.28e-06 0.00e+00 1.00e+00
#> S_2 0.00e+00 1.40e+01 1.80e+01 1.80e+01 2.20e+01 4.30e+01
#> I_2 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00
#> S_3 0.00e+00 7.50e+01 9.40e+01 9.73e+01 1.19e+02 2.06e+02
#> I_3 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00
## Run the model to generate a single stochastic trajectory.
result <- run(model)
## Summarize the trajectory. The summary includes the number of
## events by event type.
summary(result)
#> Model: SISe3
#> Number of nodes: 1600
#>
#> Transitions
#> -----------
#> S_1 -> upsilon_1*phi*S_1 -> I_1
#> I_1 -> gamma_1*I_1 -> S_1
#> S_2 -> upsilon_2*phi*S_2 -> I_2
#> I_2 -> gamma_2*I_2 -> S_2
#> S_3 -> upsilon_3*phi*S_3 -> I_3
#> I_3 -> gamma_3*I_3 -> S_3
#>
#> Global data
#> -----------
#> Parameter Value
#> upsilon_1 0.018
#> upsilon_2 0.018
#> upsilon_3 0.018
#> gamma_1 0.100
#> gamma_2 0.100
#> gamma_3 0.100
#> alpha 1.000
#> beta_t1 0.100
#> beta_t2 0.100
#> beta_t3 0.125
#> beta_t4 0.125
#> epsilon 0.000
#>
#> Local data
#> ----------
#> Parameter Value
#> end_t1 91
#> end_t2 182
#> end_t3 273
#> end_t4 365
#>
#> Scheduled events
#> ----------------
#> Exit: 182535
#> Enter: 182685
#> Internal transfer: 317081
#> External transfer: 101472
#>
#> Network summary
#> ---------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> Indegree: 40.0 57.0 62.0 62.1 68.0 90.0
#> Outdegree: 36.0 57.0 62.0 62.1 67.0 89.0
#>
#> Continuous state variables
#> --------------------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> phi 0.00e+00 0.00e+00 0.00e+00 8.92e-08 0.00e+00 1.88e-02
#>
#> Compartments
#> ------------
#> Min. 1st Qu. Median Mean 3rd Qu. Max.
#> S_1 0.00e+00 7.00e+00 9.00e+00 9.23e+00 1.20e+01 3.00e+01
#> I_1 0.00e+00 0.00e+00 0.00e+00 1.28e-06 0.00e+00 1.00e+00
#> S_2 0.00e+00 1.40e+01 1.80e+01 1.80e+01 2.20e+01 4.30e+01
#> I_2 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00
#> S_3 0.00e+00 7.50e+01 9.40e+01 9.73e+01 1.19e+02 2.06e+02
#> I_3 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00 0.00e+00